Niklas Bühler
Escaping local optima on a random walk through life.
Table of Contents
- 🎓 Academic Publications
- 🩻 Large-scale Radiograph Pre-training
- 🩻 Pilot Study for Large-scale Radiograph Pre-training
- 🧬 Functional Gene Embeddings
- 🧠 Connectome Informed Attention
- 🧊 Bayesian Deep Learning – A Stochastic Dynamics Perspective
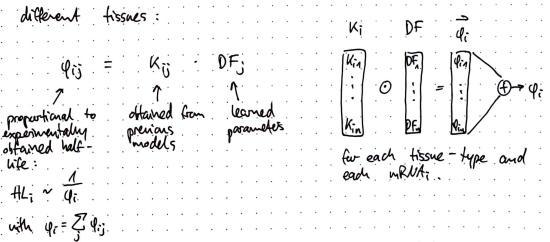
- 🧬 Interpretable Mechanistic Models for Predicting Tissue-specific RBP Expression
- 🌆 Case Study – Life Expectancy in Barcelona
- 🧮 Formalism 101
- 🗣️ Crosslingual, Language-independent Phoneme Alignment
- 🤖 Can Computers Think?
- 🔏 Security Review
- 🍛 Cooking With Curry: Lambda Poetry
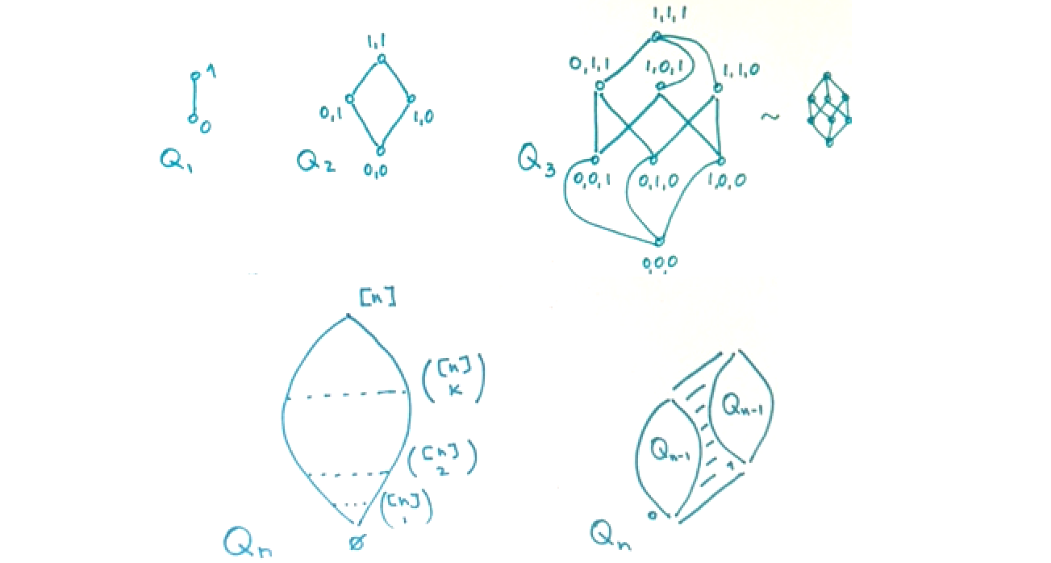
- 🌐 Graph Theory Review
- ♾️ Mächtigkeiten, Kardinalzahlen und die Kontinuumshypothese
- 🧫 Computation and Pattern Formation by Swarm Networks with Brownian Motion
🎓 Academic Publications
🩻 Large-scale Radiograph Pre-training
In recent years, medical imaging datasets have expanded significantly, offering great potential for the development of machine learning solutions in the medical field. However, manual labeling of medical data is costly and poses a significant bottleneck to their utilization.
Self-supervised learning (SSL) techniques offer a solution to this problem by extracting meaningful representations from the raw data itself, enabling label- and compute-efficient training of specialized models for downstream tasks. In this work, we demonstrate the effectiveness of SSL techniques, specifically the Masked Autoencoder (MAE) strategy, to generate such representations from a large-scale real-world clinical dataset comprising more than 600,000 radiograph images from various anatomical regions.
We introduce a novel Dynamic Batch Binning technique that reduces necessary compute by 80% when training on datasets with high image resolution variability. Our results across several clinically relevant downstream tasks show that the generated representations substantially reduce the dependence on labeled data. This is evidenced by superior performance when comparing to supervised training from scratch. Furthermore, we demonstrate the efficacy of domain-specific pre-training by achieving improved performance on the specialized medical task of fracture detection, compared to broader ImageNet-21k pre-training, despite using only 5% of its training samples and 0.5% of its training iterations during pre-training.
Our research thus demonstrates the potential of domain-specific MAE pre-training to significantly reduce the need for labeled training data in the medical domain, enabling a more effective utilization of large medical datasets with minimal labels.
🩻 Pilot Study for Large-scale Radiograph Pre-training
In recent years, medical datasets have expanded significantly, offering great potential for the development of machine learning applications in the medical field. However, manual labeling of such data is costly and poses a significant bottleneck to their utilization.
To address this issue, self-supervised learning (SSL) exploits the data itself to learn embeddings that can be quickly adapted to downstream tasks as needed.
In this work, we show the suitability of self-supervised learning techniques, specifically masked autoencoders (MAE), to generate such embeddings from a large clinical dataset comprising 12,000 radiograph images from various anatomical regions.
By pre-training a MAE model on producing these high-quality embeddings, the need for labeled data in downstream tasks is substantially reduced. This is evidenced by a linear classifier trained on representations from this MAE model achieving 84.58% top-1 accuracy on bodypart classification when using only 1% of data, marking a 7% relative improvement over fully supervised training.
This pilot study thus establishes the foundation for applying the MAE strategy to our own large-scale real-world radiograph dataset, comprising 700,000 radiograph images, as well as evaluating on more complex downstream tasks in future work.
🧬 Functional Gene Embeddings
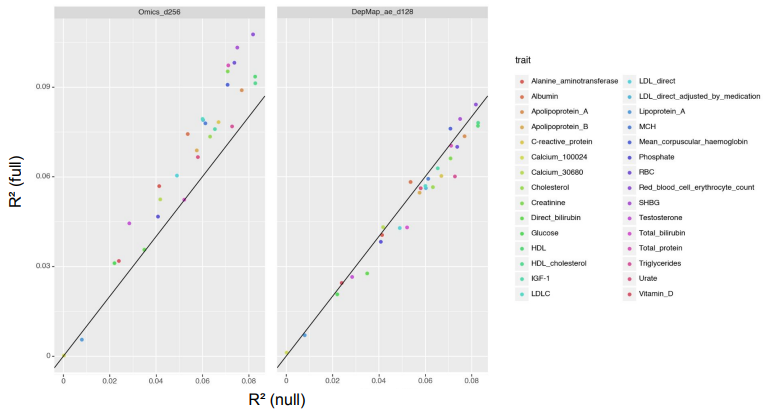
Functional gene embeddings, numerical vectors capturing gene functions, can provide useful representations of genes for downstream analyses. In this project, we extracted functional gene embeddings from transcription data and other genomewide measurements by using principle components, autoencoders and a Variational Deep Tensor Factorization model. We used these embeddings, as well as embeddings from other publications, as gene features in the prediction of genome-wide association study summary statistics for a diverse set of traits and compared their performances.
🧠 Connectome Informed Attention
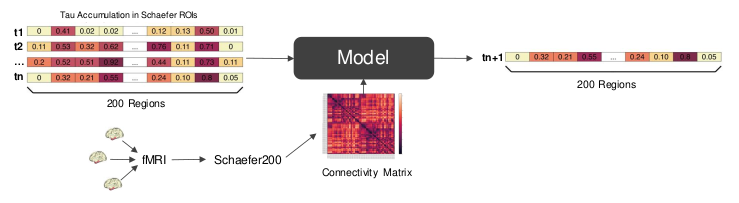
We predict tau spreading behavior for Altzheimer’s patients, based on a connectivity map and tau PET scans of the brain.
- Initial Presentation
- Second Presentation
- Third Presentation
- Fourth Presentation
- Fifth Presentation
- Sixth Presentation
- Final Poster
- Github Repository
🧊 Bayesian Deep Learning – A Stochastic Dynamics Perspective
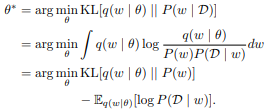
This report gives an overview of Bayesian deep learning from a stochastic dynamics perspective by first introducing Bayesian deep learning as well as two important methods for training Bayesian neural networks, and then building upon this fundament by presenting various approaches and variations inspired by stochastic dynamics.
🧬 Interpretable Mechanistic Models for Predicting Tissue-specific RBP Expression
🌆 Case Study – Life Expectancy in Barcelona
A short case study on what factors might influence life expectancy in the city of Barcelona.
🧮 Formalism 101
🗣️ Crosslingual, Language-independent Phoneme Alignment
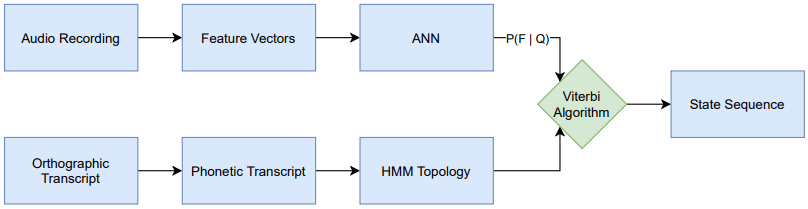
The goal of this thesis is to apply cross-lingual, multilingual techniques on the task of phoneme alignment, i.e. the task of temporally aligning a phonetic transcript to its corresponding audio recording. Three different neural network architectures are trained on a multilingual data set and utilized as a source of emission probabilities in hybrid HMM/ANN systems. These HMM/ANN systems enable the computation of phoneme alignments via the Viterbi algorithm. By iterating this process, multilingual acoustic models are bootstrapped and the resulting systems are used to cross-lingually align data from a previously unseen target language.
🤖 Can Computers Think?

Human intelligence has been the defining property of the human race for thousands of years. However, with the recent rise of machine learning, amplified by breakthrough results in deep learning, this opinion is cast in questionable light, as the opposing question starts to become more relevant and polarizing than ever before: Will computers be able to think in a way that is equivalent – or even superior – to ours?
🔏 Security Review
🍛 Cooking With Curry: Lambda Poetry
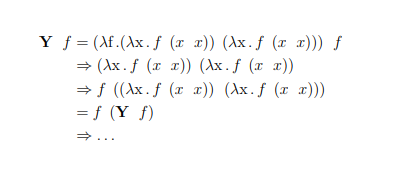
The lambda calculus is a formal system that is used to express computations. Like the Turing machine, it’s a universal model of computation, however it’s much simpler and more elegant than those bulky machines. The following pages are filled with fundamental datastructures and the most important functions operating on them, in the untyped lambda calculus. The datastructures presented differ from ordinary, imperative datastructures, as they are purely functional. That means they don’t describe where and how the data is stored, but rather how functions are applied to that data. Variable names are often chosen as a hint on the value they’re holding, but aren’t elaborated on. Their exact purpose and meaning is left open for exploration.